Overview
The Saier lab created and now maintains the Transporter Classification Database (TCDB) which was adopted by the International Union of Biochemistry and Molecular Biology (IUBMB) as the primary source of information relating to molecular transport (Saier et al, 2021). It uses a functional/phylogenetic system of classification, where each transport systems belongs to a class, subclass, family, subfamily, and system. As of March, 2021, there were 16,500 systems with >19,000 reference citations, classified into 1575 families, many of which were included in 83 superfamilies. A significant amount of our efforts involves database curation and maintenance. This is essential, given that we use the information in TCDB as the inspiration source or evaluation benchmark for many of our research projects in both the dry and wet labs. Below is a brief description of our different bioinformatic research approaches to study the evolution of transport proteins.
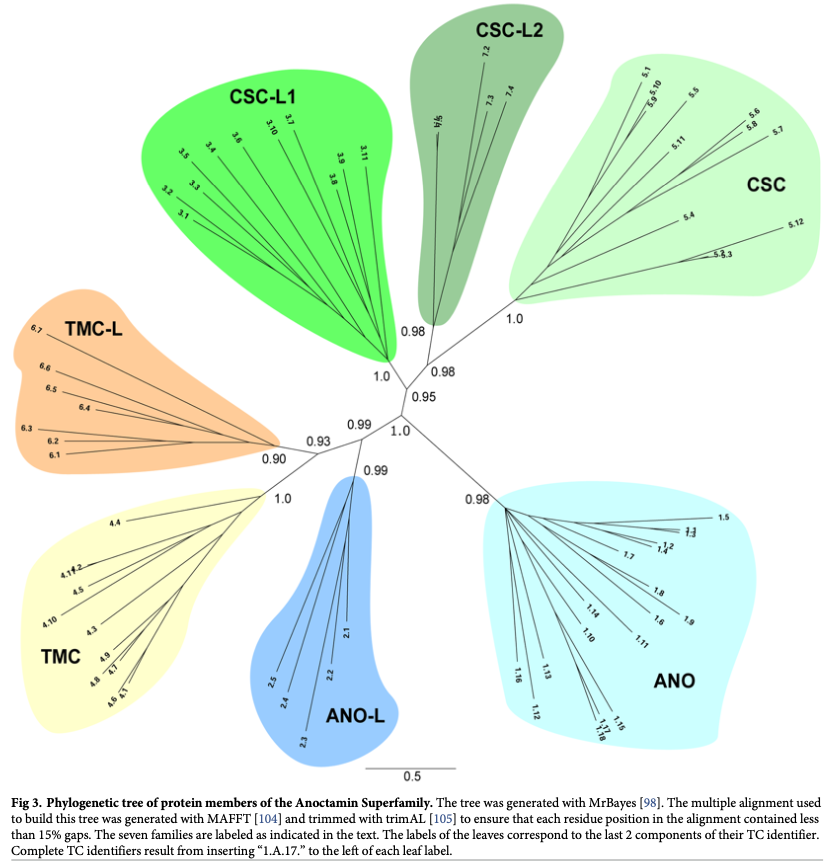
Relationships, evolution pathways, and occurrence of repeat units in transport proteins
We have shown that most transporters arose by intragenic duplication via the pathway: simple small channel proteins → large (duplicated, triplicated or quadruplicated) channel proteins → secondary carriers → primary active transporter and group translocators (Saier, 2016). Our bioinformatic research uses this information for family characterization (Reddy & Saier, 2016), and superfamily identification and expansion (Medrano-Soto et al, 2020; Wang et al, 2020).
Transportome identification and comparative genomics
We identify all transport proteins encoded within whole genomes and metagenomic sequences (i.e., the transportome), and then perform comparative analyses to study the relationships between transport systems and the physiology of the organisms. This involves identifying shared as well as unique transport systems within (meta)genomes which could give us clues as to their ecology and lifestyle (Russum et al, 2021; Zafar & Saier, 2020; Do et al, 2017).

