Find below a brief description of the three main research lines in our lab. If you want to learn more details, please click on the button “Learn More” for the specific research you are interested in and visit our Publications page.
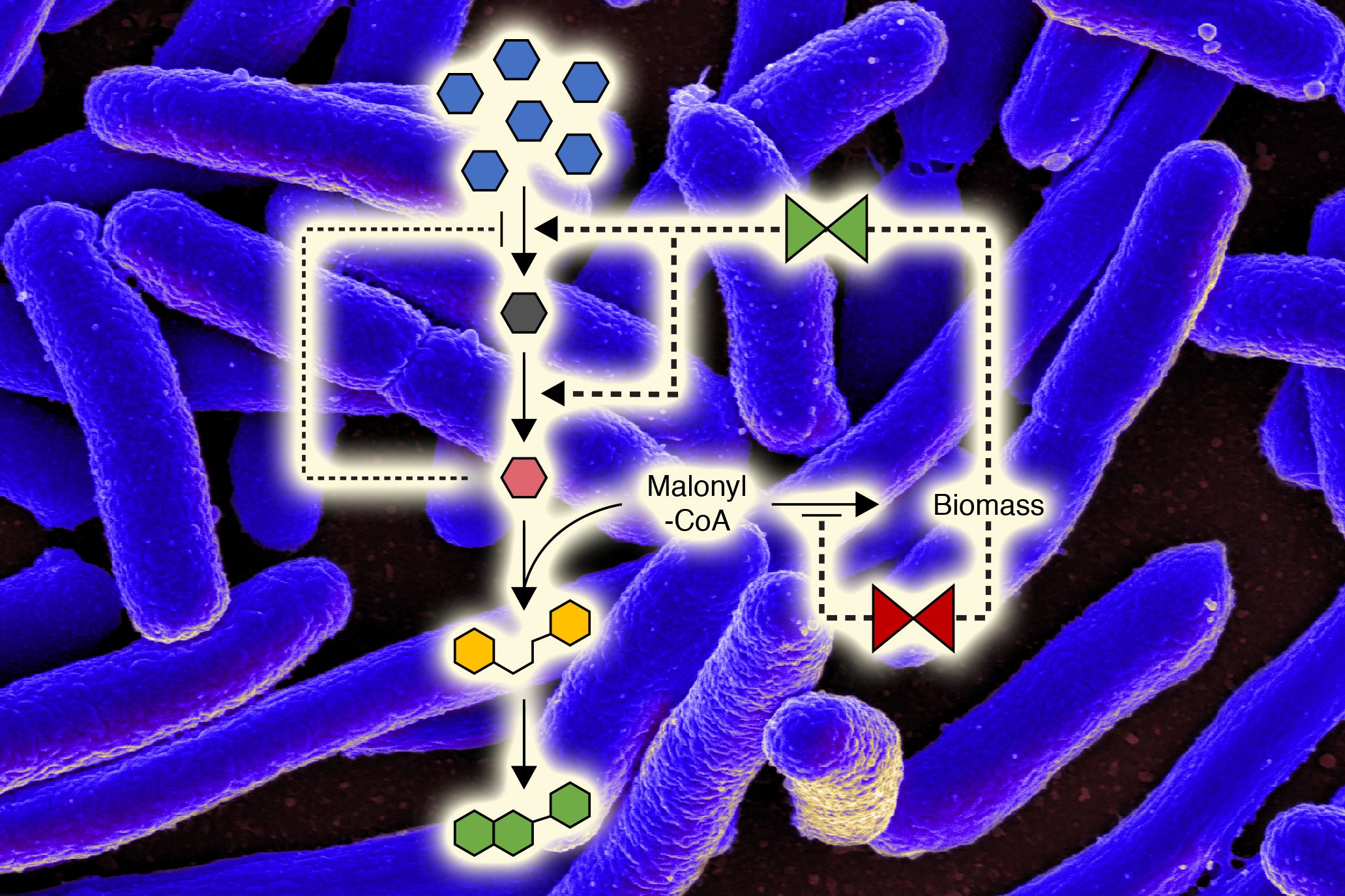
Transcriptional and metabolic regulation in bacteria
The primary pathways for cellular biosyntheses, catabolism and energy generation have been known for decades and were early evolving. They are essentially ubiquitous throughout much of the living world. However, the regulatory mechanisms controlling these pathways were late evolving, are usually phylum-specific and are generally not known, even for the best studied organism, E. coli. We have mapped the protein:protein interactions (PPIs) of all proteins in this organism (Babu et al, 2018) and then tested the biochemical and physiological consequences of these PPIs on enzyme activities…
Evolution of transport proteins
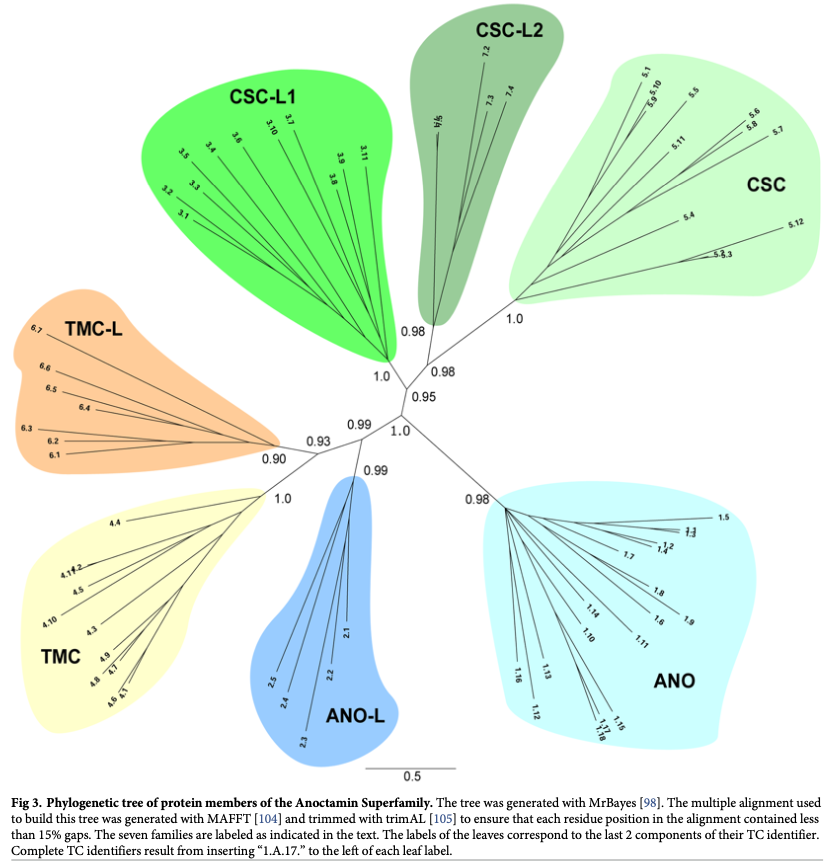
The Saier lab created and now maintains the Transporter Classification Database (TCDB) which was adopted by the International Union of Biochemistry and Molecular Biology (IUBMB) as the primary source of information relating to molecular transport (Saier et al, 2021). A significant amount of our efforts involves database curation and maintenance. This is essential, given that we use the curated information in TCDB as the inspiration source or evaluation benchmark for many of our research projects in both the dry and wet labs. Our research involves defining the relatedness, evolution pathways, and occurrence of repeat units in transport proteins…
Transposon-mediated directed mutations
Directed mutation is a proposed process that allows mutations to occur at higher frequencies when they are beneficial than when detrimental (Saier et al, 2017). Until recently, the existence of such a process has been controversial. However, we have described a novel mechanism of directed mutation mediated by the transposon, IS5 in Escherichia coli. crp deletion mutants mutate specifically to glycerol utilization (Glp+) at rates that are enhanced by glycerol or the loss of the glycerol repressor (GlpR), depressed by glucose or glpR overexpression, and RecA-independent (Zhang & Saier, 2009)…

