Overview
The primary pathways for cellular biosynthesis, catabolism and energy generation have been known for decades and were early evolving. They are essentially ubiquitous throughout much of the living world. However, the regulatory mechanisms controlling these pathways were late evolving, are usually phylum-specific and are generally not known, even for the best studied organism, E. coli. Additionally, these pathways interconnect into convoluted networks of enzymatic reactions and protein-protein interactions (PPIs). To deepen our understand on these regulatory pathways, we apply systems biology approaches, bioinformatics and in vivo experimentation on E. coli, detailing previously unknown substrate utilization pathways and their downstream effectors.
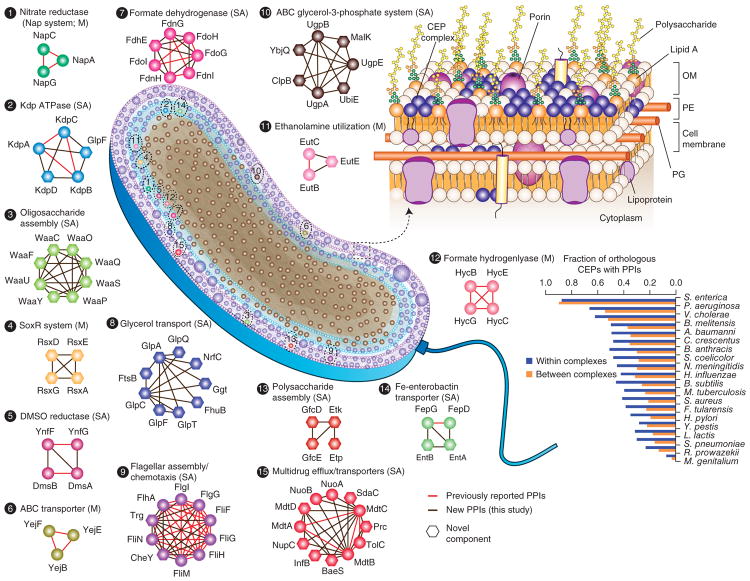
Membranes, metabolism and regulation.
We have mapped the protein-protein interactions (PPIs) of all proteins in this organism (Babu et al, 2018) and then tested the biochemical and physiological consequences of these PPIs on enzyme activities. We found for example, that many integral membrane proteins interact in the plane of the membrane to activate or inhibit their functions, depending on the interacting systems (Aboulwafa et al, 2019), and that the small phosphocarrier protein, HPr, of the PEP-dependent phosphotransferase system (PTS) is a central regulator or glycolysis, gluconeogenesis and energy balance (Rodionova et al, 2017). We also revealed that the nitrogen regulator, the PII protein, and the NanE enzyme allosterically activate glucosamine 6-phosphate deaminase, NagB, which feeds carbon from amino sugars into glycolysis and releases nitrogen (ammonium) for biosynthetic purposes. Other PPIs control enzyme cofactor activation, essential steps for the control of still other metabolic pathways (Rodionova et al, 2018). These biochemical studies are on-going and are expected to reveal that PPIs have evolved as a primary mechanism for the control of ubiquitous metabolic pathways.

