
- This event has passed.
Master’s thesis defense by Sophia Beliaev
At 2pm on March 15, 2024, our grad student Sophia Beliaev will defend her master’s thesis entitled “Expansion of the Major Facilitator Superfamily (MFS) with the Sar TMS10 Membrane Protein (S10MP) Family.” The zoom link will be provided before the defense.
Abstract
Transport proteins are crucial to essential cellular processes. Databases such as the Transporter Classification Database (TCDB) hold a wealth of hypothetical transport proteins in need of adequate characterization. However, there are significant challenges to transport protein experimental research in vitro and in vivo due to the physico-chemical properties of transport proteins linked to their membrane-bound nature. Instead, bioinformatic methods may be utilized to elucidate structural features, which facilitate the identification of homology between families. If one of the families is well characterized, it may provide clues to the potential functions and mechanisms of action of the poorly understood family.In this project, potential homologous relationships were explored between the well-characterized Major Facilitator Superfamily (MFS) and two uncharacterized transport protein families: the Large 14-16 Putative Transporter (LPT) Family and the Sar TMS10 Membrane Protein (S10MP) Family. To infer homology between the families, we searched for seven types of bioinformatic evidence: 1) sequence similarity, 2) topology and hydropathy profile compatibility, 3) shared domains, 4) conserved motifs, 5) similarity of hidden Markov model-profiles at the family level, 6) common 3D structural folds, and 7) a superfamily tree supporting the proposed relationships. Our results showed that one established MFS family, The Anion: Cation Symporter (ACS) Family, generated reliable signals of homology with the S10MP family. Therefore, we concluded that the S10MP family is a distant member of the MFS. Characteristics of the MFS proteins may extend to S10MP members, warranting further research.
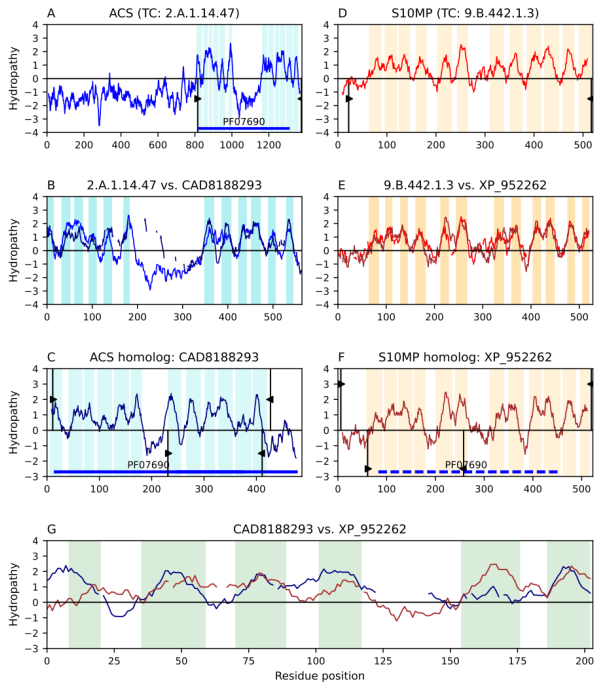
Figure 1. Transitivity path of homology between the second repeat unit (RU) of ACS and the first RU of S10MP. Hydropathy plots are presented across the homology transitivity path for both families. The x-axis corresponds to amino acid residues in the protein sequences and the y-axis indicates hydropathy. Panels A-C depict relationships within the ACS family, panels D-F depict relationships within the S10MP family, and panel G presents the evidence supporting the relationship between the two families. Cyan (ACS) and orange (S10MP) bars denote TMSs. Thing vertical black lines with wedges delimit the region of a protein involved in an alignment. Wedges with positive hydropathy values indicate the sequence region involved in the sequence alignment displayed in the panel above. Similarly, black wedges with negative hydropathy values indicate the sequence regions involved in the sequence alignment displayed in the panel below. Interruptions in the hydropathy plots of panels B, E, and G indicate gaps in the corresponding sequence alignments. Colored solid horizontal lines depict domains with direct Pfam hits in the annotated protein. Colored dashed horizontal lines depict domains projected onto the protein from the family which had a direct hit with the Pfam domain. A) Hydropathy plots of ACS member Q8I2Z2 (TC: 2.A.1.14.47). B) Hydropathy plot of the alignment (E-value: 4.7e-24) between the ACS member and its homolog. C) Hydropathy plot of ACS homolog CAD8188293. D) Hydropathy plots of S10MP member XP_021337925 (TC: 9.B.442.1.3). E) Hydropathy plots of the alignment (E-value: 9.2e-31) between the S10MP member and its homolog. F. Hydropathy plot of S10MP homolog XP_952262. G) Hydropathy plot of the alignment (E-value: 2.3e-3) between ACS and S10MP homologs. Pfam domain PF07690 in family ACS was projected to the S10MP homolog XP_952262 (E-value: 0.0027; see Methods).

