Abstract
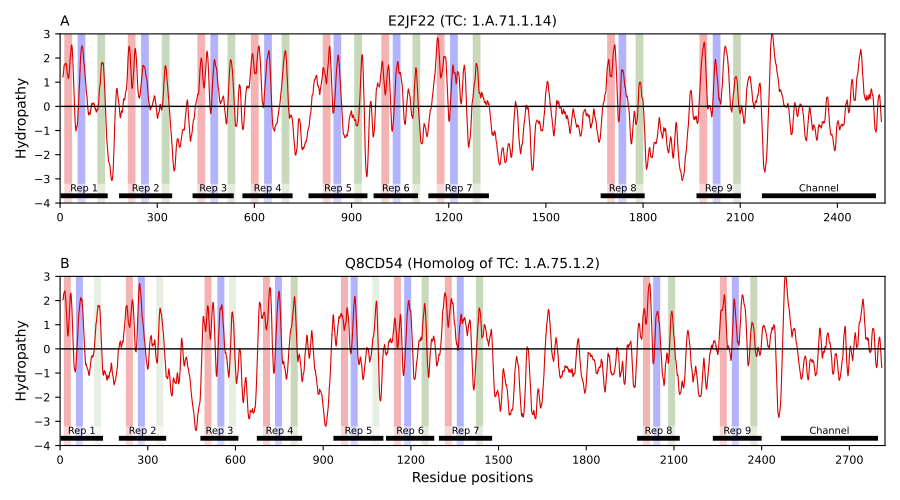
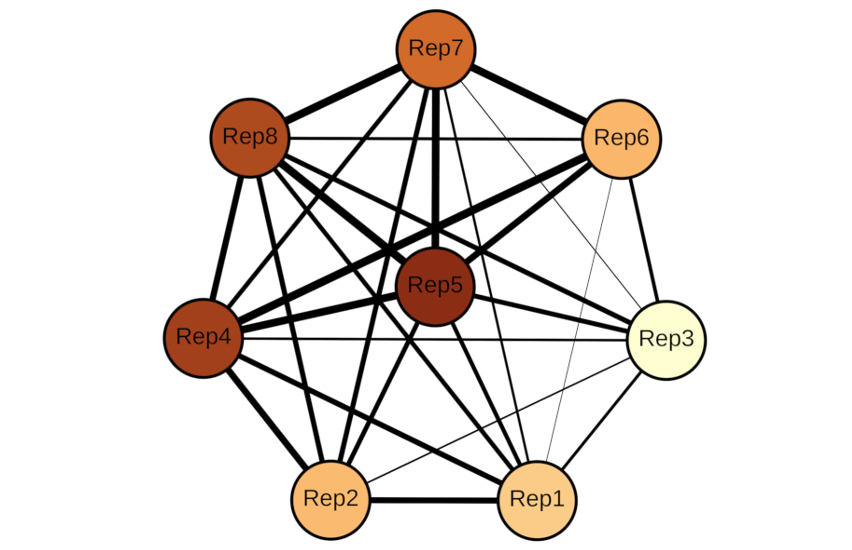
Members of the Piezo family of mechanically activated cation channels are involved in multiple physiological processes including vascular development, cell differentiation, touch perception, hearing and more. Mutations in these proteins are associated with a variety of diseases, such as colorectal adenomatous polyposis, dehydrated hereditary stomacytosis, and hereditary xerocytosis. Available 3D structures for Piezo proteins show nine regions of four transmembrane segments each that have the same fold. Despite the remarkable similarity among the nine characteristic structural repeats in the family, no significant sequence similarity among them has been reported. Using a bioinformatics approach and the Transporter Classification Database (TCDB) as reference, we reliably identified sequence similarity among repeats based on four lines of evidence: (1) HMM-profile similarities across repeats at the family level, (2) pairwise sequence similarities between different repeats across Piezo homologs, (3) Piezo-specific conserved sequence signatures that consistently identify the same regions across repeats, and (4) conserved residues that maintain the same orientation and location in 3D space.