MSc Daniel Tyler, grad student Kevin Hendargo, Dr. Medrano-Soto and Dr. Saier just published a paper in the journal Microbial Physiology. The title is: Discovery and Characterization of the Phospholemman/SIMP/Viroporin Superfamily. For your convenience, this is the link to PubMed.
Abstract
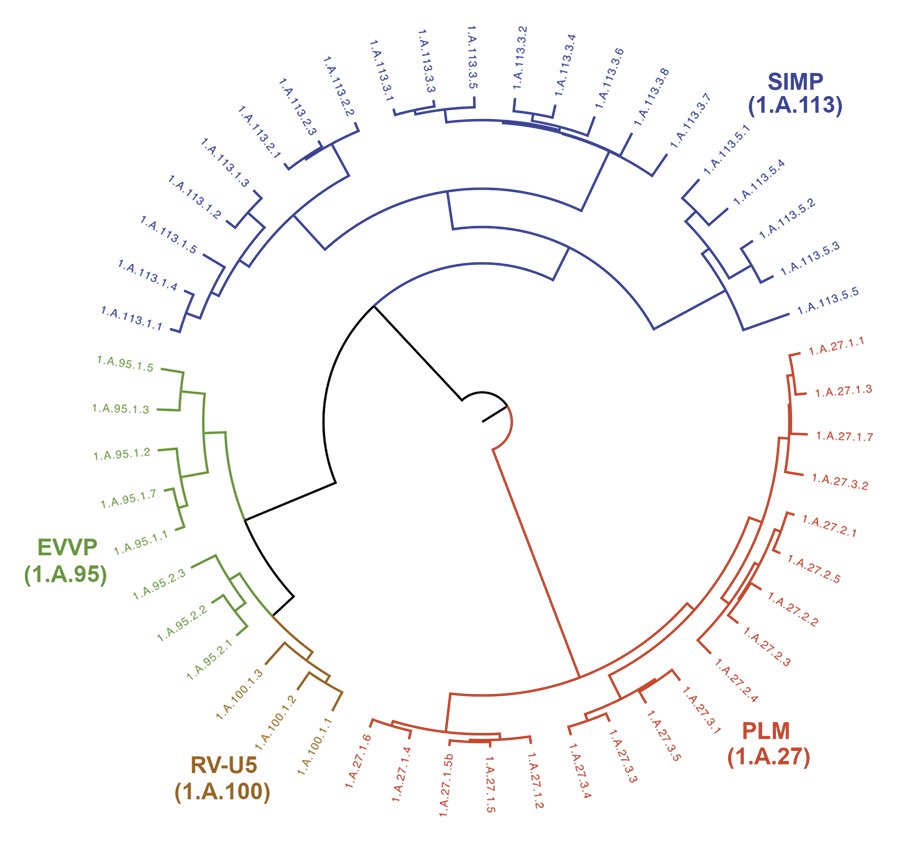
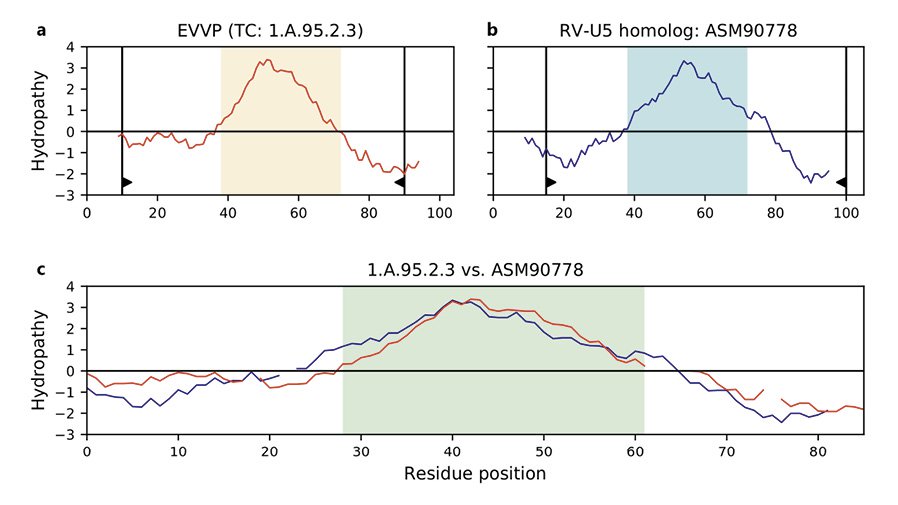
Using bioinformatic approaches, we present evidence of distant relatedness among the Ephemerovirus Viroporin family, the Rhabdoviridae Putative Viroporin U5 family, the Phospholemman family, and the Small Integral Membrane Protein family. Our approach is based on the transitivity property of homology complemented with five validation criteria: (1) significant sequence similarity and alignment coverage, (2) compatibility of topology of transmembrane segments, (3) overlap of hydropathy profiles, (4) conservation of protein domains, and (5) conservation of sequence motifs. Our results indicate that Pfam protein domains PF02038 and PF15831 can be found in or projected onto members of all four families. In addition, we identified a 26-residue motif conserved across the superfamily. This motif is characterized by hydrophobic residues that help anchor the protein to the membrane and charged residues that constitute phosphorylation sites. In addition, all members of the four families with annotated function are either responsible for or affect the transport of ions into and/or out of the cell. Taken together, these results justify the creation of the novel Phospholemman/SIMP/Viroporin superfamily. Given that transport proteins can be found not just in cells, but also in viruses, the ability to relate viroporin protein families with their eukaryotic and bacterial counterparts is an important development in this superfamily.