Congratulations to our grad student Daniel Tyler for a successful defense of his Master’s thesis on Jan 28, 2021. The title of his thesis is “Discovery and Characterization of the Phospholemman/SIMP/Viroporin (PSV) Superfamily”.
Abstract
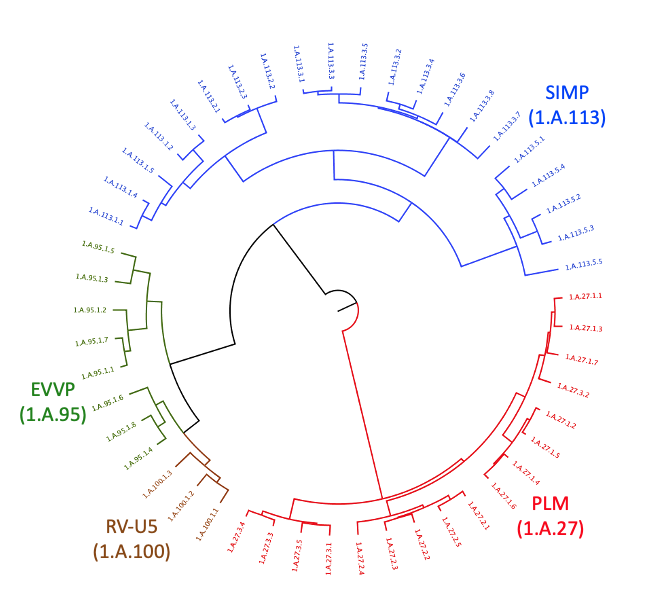
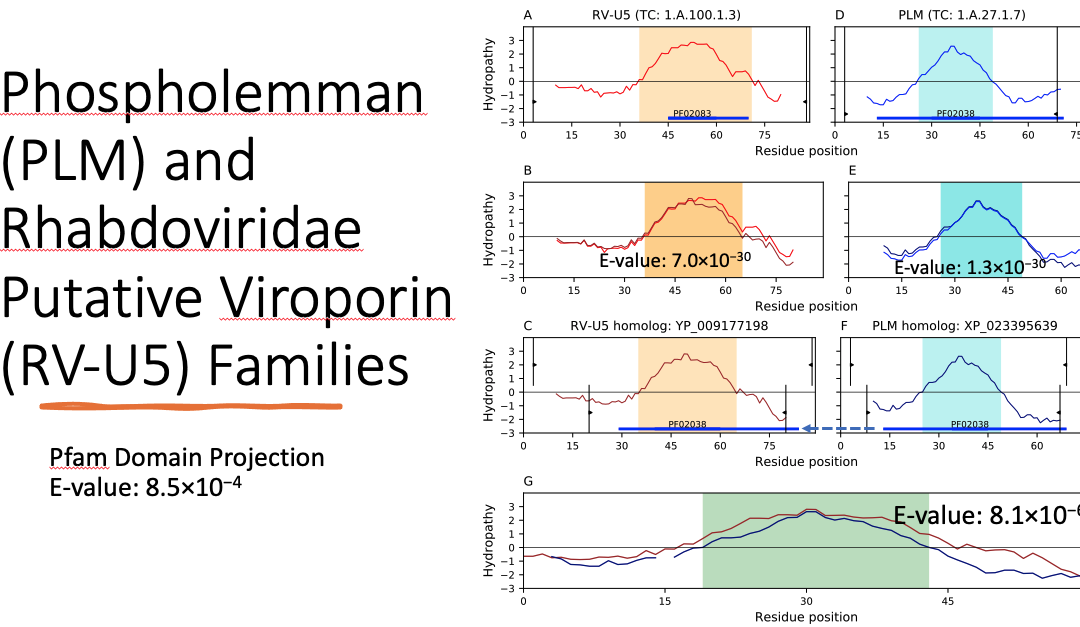
Determining the relatedness between biological entities can translate to knowledge which would otherwise be difficult to uncover without greater context. In particular, identifying homology between proteins often provides insight on previous unknowns such as structure, function, and even substrate identity. Given that transport proteins can be found not just in cells, but also in viruses, the ability to relate viroporin protein families with their eukaryotic and bacterial counterparts is an exciting development in superfamily formation. Using a number of bioinformatic techniques to compare the Ephemerovirus Viroporin (EVVP) family, the Rhabdoviridae Putative Viroporin (RV-U5) family, the Phospholemman (PLM) family, and the Small Integral Membrane Protein (SIMP) family, we examined the protein families as candidates for a superfamily on the basis of protein sequence similarity, compatibility of hydropathy profiles, topology of transmembrane segments (TMSs) and conservation of protein domains and sequence motifs. Our results indicate that the Pfam domains ATP1G1_PLM_MAT8 (PF02038) and DUF4713 (PF15831) can be found in or projected onto all four families. In addition, we identified a 26-residue motif conserved across the superfamily which is located within the PF02038 domain. This motif is characterized by hydrophobic residues that help anchor the protein to the membrane and charged residues that constitute phosphorylation sites. Taken together, these pieces of evidence justify the creation of the novel Phospholemman/SIMP/Viroporin (PSV) Superfamily.